
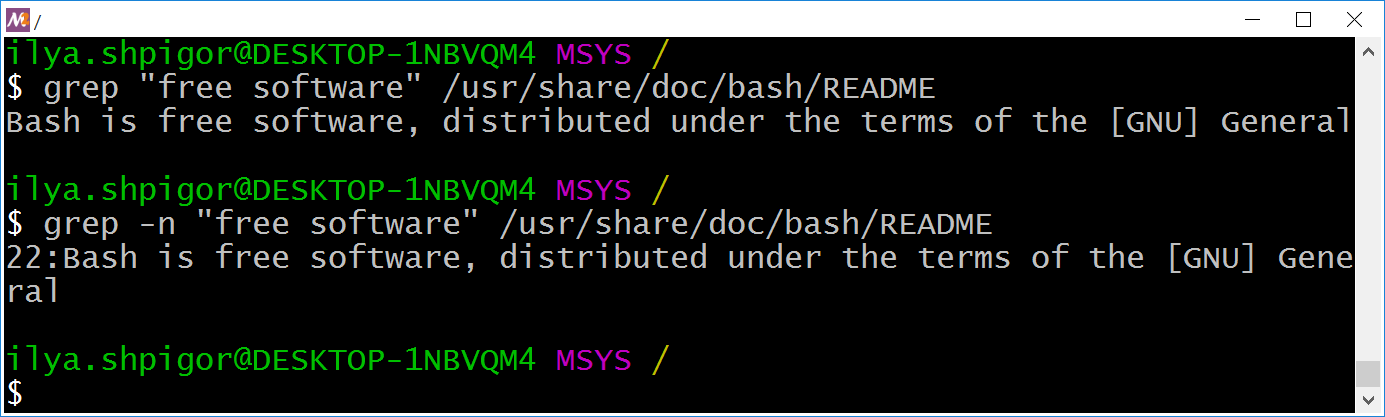
> zcat reads.fq.gz | awk '/ATGATGATG/ ' reads.fastq If the file is in FASTA format, we will count the number of sequences like this: > grep -c "^>" reads.faĪlso we can count how many times appear an specific subsequence: > zgrep -c 'ATGATGATG' reads.fq.gz If we want to check the contents of the file we can use the command ‘less’ or ‘zless’: > less reads.fq.gzĪnd to count the number of sequences stored into the file we can count the number of lines and divide by 4: > zcat reads.fq.gz | echo $((`wc -l`/4)) The resulting file will be named ‘reads.fq.gz’ by default. To start, let’s compress a FASTQ file in GZIP format: > gzip reads.fq In the next lines I’ll show you some commands to deal with compressed FASTQ files, with minor changes they also can be used with uncompressed ones and FASTA format files. When these files are compressed with GZIP their sizes are reduced in more than 10 times (ZIP format is less efficient). A FASTQ file usually contain millions of sequences and takes up dozens of Gigabytes in a disk. Here I’ll summarize some Linux commands that can help us to work with millions of DNA sequences from New Generation Sequencing (NGS).Ī file storing biological sequences with extension ‘.fastq’ or ‘.fq’ is a file in FASTQ format, if it is also compressed with GZIP the suffix will be ‘.fastq.gz’ or ‘.fq.gz’.


 0 kommentar(er)
0 kommentar(er)
